Advanced Topics
Prerequisites
- 0. Before You Start
- 2. Installation
- 3. Hello, World!
- 4. Run a Real Simulation
- Python virtual environment containing the following packages
- merlin
- pandas
- faker
Estimated Time
15 minutes
You Will Learn
- Run workflows using HPC batch schedulers
- Distribute workflows across multiple batch allocations and machines
- Setup iterative workflow specs suited for optimization and dynamic sampling applications
Setup
The code for the following examples can be obtained from command line, invoking:
This will copy the three Merlin workflow specifications from this section and the supporting python scripts. Each specification may need some modification to adapt it to the batch scheduler you will be using. In addition, the dynamic sampling workflow will need an additional modification to set the path of the virtual environment, which is set as a variable in the env block.
Interfacing with HPC Systems
Another block is added to the Merlin workflow specification when running on HPC systems—the batch block. This block contains information about the batch scheduler system, such as batch type, batch queue to use, and banks to charge. There are additional optional arguments for addressing any special configurations or launch command arguments, varying based on batch type. In addition, the shell type used by each step's cmd scripts can be specified here. While the number of nodes in a batch allocation can be defined here, it will be overridden in the worker config.
batch:
# Required keys:
type: flux
bank: testbank
queue: pbatch
# Optional keys:
flux_path: <optional path to flux bin>
flux_start_opts: <optional flux start options>
flux_exec_workers: <optional, flux argument to launch workers on
all nodes. (True)>
launch_pre: <Any configuration needed before the srun or jsrun launch>s
launch_args: <Optional extra arguments for the parallel launch command>
worker_launch: <Override the parallel launch defined in merlin>
shell: <the interpreter to use for the script after the shebang>
# e.g. /bin/bash, /bin/tcsh, python, /usr/bin/env perl, etc.
nodes: <num nodes> # The number of nodes to use for all workers
This can be overridden in the workers config.
If this is unset the number of nodes will be
queried from the environment, failing that, the
number of nodes will be set to 1.
Inside the study step specifications are a few additional keys that become more useful on HPC systems: nodes, procs, and task_queue. Adding on the actual study steps to the above batch block specifies the actual resources each steps processes will take.
study:
- name: sim-runs
description: Run simulations
run:
cmd: $(LAUNCHER) echo "$(VAR1) $(VAR2)" > simrun.out
nodes: 4
procs: 144
task_queue: sim_queue
- name: post-process
description: Post-Process simulations on second allocation
run:
cmd: |
cd $(runs1.workspace)/$(MERLIN_SAMPLE_PATH)
$(LAUNCHER) <parallel-post-proc-script>
nodes: 1
procs: 36
depends: [sim-runs]
task_queue: post_proc_queue
In addition to the batch block is the resources section inside the merlin block. This can be used to put together custom celery workers. Here you can override batch types and node counts on a per worker basis to accommodate steps with different resource requirements. In addition, this is where the task_queue becomes useful, as it groups the different allocation types, which can be assigned to each worker here by specifying step names.
merlin:
resources:
task_server: celery
# Flag to determine if multiple workers can pull tasks
# from overlapping queues. (default = False)
overlap: False
# Customize workers. Workers can have any user-defined name
# (e.g., simworkers, learnworkers, ...)
workers:
simworkers:
args: <celery worker args> # <optional>
steps: [sim-runs] # <optional> [all] if none specified
nodes: 4 # optional
machines: [host1] # <optional>
Arguments to celery itself can also be defined here with the args key. Of particular interest will be:
|
|
num_threads |
|
|
num_tasks |
|
|
Concurrency can be used to run multiple workers in an allocation, thus is recommended to be set to the number of simulations or step work items that fit into the number of nodes in the batch allocation in which these workers are spawned.
Note
Some schedulers, such as flux, can support more jobs than the node has resources for. This may not impact the throughput, but it can prevent over-subscription errors that might otherwise stop the workflow.
The prefetch multiplier is more related to packing in tasks into the time of the allocation. For long running tasks it is recommended to set this to 1. For short running tasks, this can reduce overhead from talking to the rabbit servers by requesting <num_threads> x <num_tasks> tasks at a time from the server.
The -0 fair option enables workers running tasks from different queues to run on the same allocation.
The example block below extends the previous with workers configured for long running simulation jobs as well as shorter running post processing tasks that can cohabit an allocation:
merlin:
resources:
task_server: celery
overlap: False
# Customize workers
workers:
simworkers:
args: --concurrency 1
steps: [sim-runs]
nodes: 4
machines: [host1]
postworkers:
args: --concurrency 4 --prefetch-multiplier 2
steps: [post-proc-runs]
nodes: 1
machines: [host1]
Putting it all together and adding some variables/samples we have an HPC batch enabled study specification. In this demo workflow, sample_names generates many single core jobs, with concurrency set to 36 for this particular machine that has 36 cores per node. The collect step on the other hand consists of a single job that uses all cores on the node, and is assigned to a queue that has a concurrency of 1.
HPC Batch Enabled Study Spec
description:
name: hpc_demo
description: Demo running a workflow on HPC machines
env:
variables:
OUTPUT_PATH: ./name_studies
# Collect individual sample files into one for further processing
COLLECT: $(SPECROOT)/sample_collector.py
# Process single iterations' results
POST_PROC: $(SPECROOT)/sample_processor.py
# Process all iterations
CUM_POST_PROC: $(SPECROOT)/cumulative_sample_processor.py
# Number of threads for post proc scripts
POST_NPROCS: 36
PYTHON: <INSERT PATH TO VIRTUALENV HERE>
batch:
type: flux
bank: testbank
queue: pdebug
shell: /bin/bash
nodes: 1
########################################
# Study definition
########################################
study:
- name: sample_names
description: Record samples from the random name generator
run:
cmd: |
$(LAUNCHER) echo "$(NAME)"
$(LAUNCHER) echo "$(NAME)" > name_sample.out
nodes: 1
procs: 1
task_queue: name_queue
- name: collect
description: Collect all samples generated
run:
cmd: |
echo $(MERLIN_GLOB_PATH)
echo $(sample_names.workspace)
ls $(sample_names.workspace)/$(MERLIN_GLOB_PATH)/name_sample.out | xargs $(PYTHON) $(COLLECT) -out collected_samples.txt --np $(POST_NPROCS)
nodes: 1
procs: 1
depends: [sample_names_*]
task_queue: post_proc_queue
- name: post-process
description: Post-Process collection of samples, counting occurrences of unique names
run:
cmd: |
$(PYTHON) $(POST_PROC) $(collect.workspace)/collected_samples.txt --results iter_$(ITER)_results.json
nodes: 1
procs: 1
depends: [collect]
task_queue: post_proc_queue
########################################
# Worker and sample configuration
########################################
merlin:
resources:
task_server: celery
overlap: False
workers:
nameworkers:
args: --concurrency 36 --prefetch-multiplier 3
steps: [sample_names]
nodes: 1
machines: [quartz]
postworkers:
args: --concurrency 1 --prefetch-multiplier 1
steps: [post-process]
nodes: 1
machines: [quartz]
###################################################
samples:
column_labels: [NAME]
file: $(MERLIN_INFO)/samples.csv
generate:
cmd: |
$(PYTHON) $(SPECROOT)/faker_sample.py -n 200 -outfile=$(MERLIN_INFO)/samples.csv
Multi-Machine Workflows
Spreading this workflow across multiple machines requires a small modification of the above workflow: simply add additional host names to the machines list in the worker config. A caveat for this feature is that all host systems will need to have access to the same workspace/filesystem.
The following resource block demonstrates usage of one host for larger simulation steps, and a second host for the smaller post processing steps. In this case you simply need to get an allocation on each host, then execute run-workers on each. The run command only needs to be run one time from any host to send the tasks to the central task queue server.
########################################
# Worker and sample configuration
########################################
merlin:
resources:
task_server: celery
overlap: False
# Customize workers
workers:
simworkers:
args: --concurrency 1
steps: [sim-runs]
nodes: 4
machines: [host1]
postworkers:
args: --concurrency 4 --prefetch-multiplier 2
steps: [post-proc-runs]
nodes: 1
machines: [host2]
Dynamic Task Queuing and Sampling
Iterative workflows, such as optimization or machine learning, can be implemented in Merlin via recursive workflow specifications that use dynamic task queuing. The example spec below is a simple implementation of this using an iteration counter $(ITER) and a predetermined limit $(MAX_ITER) to limit the number of times to generate new samples and spawn a new instantiation of the workflow. The iteration counter takes advantage of the ability to override workflow variables on the command line.
Iterative Spec
description:
name: dynamic_sampling_demo
description: Demo dynamic sampling workflow
env:
variables:
OUTPUT_PATH: ./name_studies
ITER_OUTPUT: $(SPECROOT)/$(OUTPUT_PATH)/iter_outputs # Iteration and cumulative results
COLLECT: $(SPECROOT)/sample_collector.py
POST_PROC: $(SPECROOT)/sample_processor.py # Process single iterations' results
CUM_POST_PROC: $(SPECROOT)/cumulative_sample_processor.py # Process all iterations
POST_NPROCS: 36 # Number of threads for post proc scripts
PYTHON: <INSERT PATH TO VIRTUALENV HERE>
ITER: 1
MAX_ITER: 10
batch:
type: flux
bank: testbank
queue: pdebug
shell: /bin/bash
nodes: 1
########################################
# Study definition
########################################
study:
- name: sample_names
description: Record samples from the random name generator
run:
cmd: |
$(LAUNCHER) echo "$(NAME)"
$(LAUNCHER) echo "$(NAME)" > name_sample.out
nodes: 1
procs: 1
task_queue: name_queue
- name: collect
description: Collect all samples generated
run:
cmd: |
echo $(MERLIN_GLOB_PATH)
echo $(sample_names.workspace)
ls $(sample_names.workspace)/$(MERLIN_GLOB_PATH)/name_sample.out | xargs $(PYTHON) $(COLLECT) -out collected_samples.txt --np $(POST_NPROCS)
nodes: 1
procs: 1
depends: [sample_names_*]
task_queue: post_proc_queue
- name: post-process
description: Post-Process collection of samples, counting occurrences of unique names
run:
cmd: |
$(PYTHON) $(POST_PROC) $(collect.workspace)/collected_samples.txt --results $(ITER_OUTPUT)/iter_$(ITER)_results.json
nodes: 1
procs: 1
depends: [collect]
task_queue: post_proc_queue
- name: run-more-samples
description: Generate new set of samples and rerun, or generate some descriptive plots/statistics
run:
cmd: |
if [ $(ITER) -ge $(MAX_ITER) ] ; then
echo "done"
$(PYTHON) $(CUM_POST_PROC) $(ITER_OUTPUT)/iter_*_results.json --np $(POST_NPROCS) --hardcopy $(ITER_OUTPUT)/cumulative_results.png
else
next_iter=$(ITER)
((next_iter=next_iter+1))
echo "Starting iteration " $next_iter
cd $(SPECROOT)
merlin run $(SPECROOT)/faker_demo.yaml --vars ITER=$next_iter
fi
nodes: 1
procs: 1
depends: [post-process]
task_queue: post_proc_queue
########################################
# Worker and sample configuration
########################################
merlin:
resources:
task_server: celery
overlap: False
# Customize workers NOTE: abuse this for scaling study: prefetch mult increase
# - celery->rabbit query overhead for fast jobs
workers:
nameworkers:
args: --concurrency 36 --prefetch-multiplier 3
steps: [sample_names]
nodes: 1
machines: [borax, quartz]
# NOTE: specifying wrong step leaves orphaned queue -> purge first!
# also, invalid host name appears to fail silently
postworkers:
args: --concurrency 1 --prefetch-multiplier 1
steps: [post-process]
nodes: 1
machines: [borax, quartz]
###################################################
samples:
column_labels: [NAME]
file: $(MERLIN_INFO)/samples.csv
generate:
cmd: |
$(PYTHON) $(SPECROOT)/faker_sample.py -n 200 -outfile=$(MERLIN_INFO)/samples.csv
This workflow specification is intended to be invoked within an allocation of nodes on your HPC cluster, e.g. within and sxterm. The last step is to queue up new samples for the next iteration, merlin run faker_demo.yaml .... Notice how we don't need another run-workers call since the workers from the first instantiation are still alive. Thus the new samples will immediately start processing on the existing allocation.
Another change in this workflow relative to the single stage version is managing the workspaces and outputs. The strategy used here is to create a new directory for collecting each iterations final outputs, $(ITER_OUTPUT), facilitating collective post processing at the end without having to worry about traversing into each iterations' local workspaces.
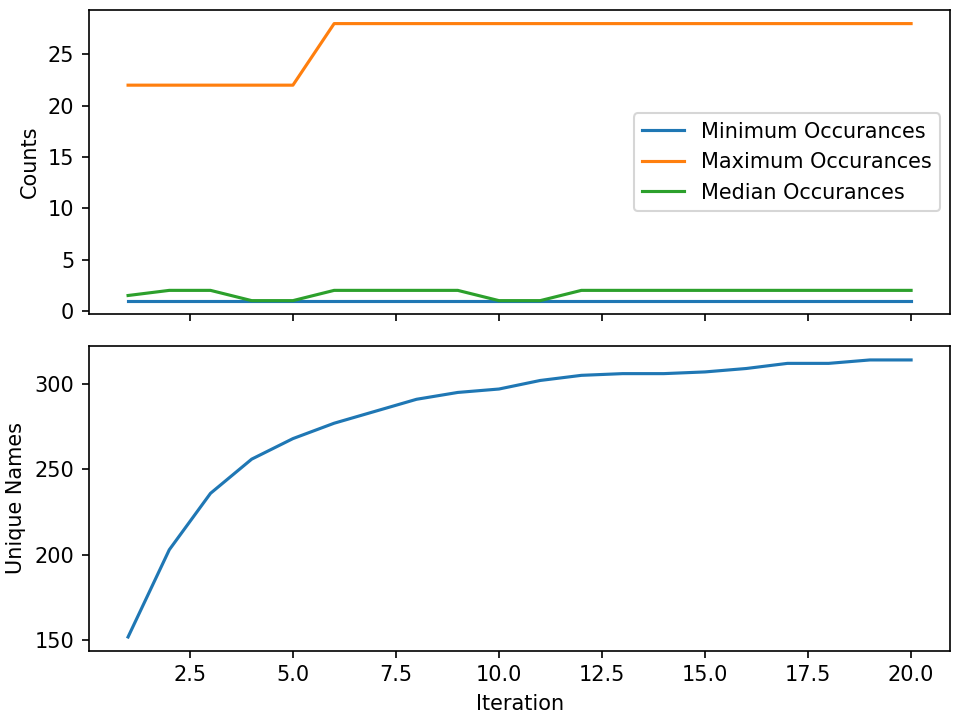
The workflow itself isn't doing anything practical; it's simply repeatedly sampling from a fake name generator in an attempt to count the number of unique names that are possible. The figure below shows results from running 20 iterations, with the number of unique names faker can generate appearing to be slightly more than 300.